07: Combining data from multiple netcdf files#
In some cases, people will choose to break down their data into multiple smaller netcdf files that they will publish in a single data collection. There are a number of good reasons to do this.
The data user can access only the data they are interested in.
Each file can be simpler with potentially less dimensions and less missing values. Imagine you have 10 depth profiles that all sample a different set of depths. If these profiles were included in a single netcdf file, the file would most likely a single depth dimension and coordinate variable which would need to account for all 10 depth profiles. Alternatively, 10 depth dimensions and coordinate variables could be included.
Each individual file can be assigned a separate set of global attributes which describe the data more accurately. For example each file could have global attributes for the coordinates and timestamp. If multiple depth profiles are stored in a single file, only the minimum and maximum coordinates and timestamp can be encoded into the global attributes.
Imagine you are looking for data in a data centre. You want to find depth profiles in a certain area of interest on a map. Files that include a single depth profile will be presented as points on the map. Files that include multiple depth profiles will be presented as a bounding box on a map, and without opening up the file it could be unclear whether the file includes data for your area of interest.
A common reaction to learning that data are divided into multiple files is that extracting the data will involve more work. However, if the files are similar (and they should be if they follow the CF and ACDD conventions) this is not neccessarily the case.
In this notebook we will look at how to combine data from multiple netcdf files into a single object (e.g. dataframe, multi-dimensional array) that you can use.
from IPython.display import YouTubeVideo
YouTubeVideo('AbLRV5YUW2g')
Introducing the data#
The link below is an OPeNDAP access point to CTD data collected as part of the Nansen Legacy data. The data are grouped together by cruise and each CF-NetCDF file contains data from a single depth profile. https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/
Each file has it’s own access point like this https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708/CTD_station_ISG_SVR1_-Nansen_Legacy_Cruise-_2021_Joint_Cruise_2-1.nc.html
And anyone can download some or all of the data from the CF-NetCDF file into ASCII files. This makes data access a lot easier for people who don’t yet know how to work with NetCDF files.
We can load into Python by removing the .html suffix and including the rest of the URL as our filepath. You don’t have to download the data!
import xarray as xr
xrds = xr.open_dataset('https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708/CTD_station_ISG_SVR1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc')
xrds
<xarray.Dataset>
Dimensions: (PRES: 259)
Coordinates:
* PRES (PRES) float32 2.0 3.0 4.0 5.0 ... 257.0 258.0 259.0 260.0
Data variables: (12/33)
PRES_QC (PRES) float32 ...
TEMP (PRES) float32 ...
PSAL (PRES) float32 ...
FLU2 (PRES) float32 ...
CNDC (PRES) float32 ...
DENS (PRES) float32 ...
... ...
OXYOCPVL-1_QC (PRES) float32 ...
SPAR_QC (PRES) float32 ...
PAR_QC (PRES) float32 ...
PSAL-2_QC (PRES) float32 ...
TEMP-2_QC (PRES) float32 ...
ATTNZS01_QC (PRES) float32 ...
Attributes: (12/73)
qc_manual: Recommendations for in-situ data Near Re...
contact: datahjelp@hi.no
distribution_statement: These data are public and free of charge...
naming_authority: no.unis
license: https://creativecommons.org/licenses/by/...
data_assembly_center: IMR
... ...
geospatial_vertical_resolution: 1 dbar
pi_email: jannes@unis.no
pi_institution: University Centre in Svalbard
station_name: ISG/SVR1
metadata_link: https://doi.org/10.21335/NMDC-2085836005...
_NCProperties: version=2,netcdf=4.6.3,hdf5=1.10.5Let’s look at a quick example of how we can extract the data into numpy arrays
temperature = xrds['TEMP'].values
salinity = xrds['PSAL'].values
temperature, salinity
(array([3.226, 3.211, 3.202, 3.205, 3.213, 3.215, 3.21 , 3.215, 3.191,
3.162, 3.144, 3.118, 3.081, 2.993, 2.913, 2.861, 2.837, 2.826,
2.809, 2.776, 2.743, 2.703, 2.687, 2.652, 2.587, 2.544, 2.508,
2.483, 2.472, 2.434, 2.399, 2.355, 2.298, 2.277, 2.252, 2.225,
2.201, 2.173, 2.113, 2.073, 2.06 , 2.04 , 2.022, 2.006, 2.004,
2.008, 2.067, 2.126, 2.145, 2.143, 2.248, 2.288, 2.232, 2.212,
2.136, 1.997, 1.748, 1.594, 1.657, 1.623, 1.468, 1.385, 1.374,
1.383, 1.378, 1.38 , 1.367, 1.351, 1.38 , 1.376, 1.391, 1.396,
1.403, 1.398, 1.413, 1.393, 1.371, 1.248, 1.159, 1.063, 0.997,
0.981, 0.944, 0.933, 0.928, 0.964, 1. , 1.013, 1.044, 1.095,
1.241, 1.216, 1.154, 1.281, 1.362, 1.4 , 1.378, 1.353, 1.237,
1.069, 0.993, 0.989, 1.048, 1.078, 1.07 , 1.05 , 1.016, 0.946,
0.928, 0.984, 1.004, 0.942, 0.892, 0.828, 0.691, 0.641, 0.635,
0.632, 0.625, 0.615, 0.566, 0.542, 0.518, 0.493, 0.52 , 0.457,
0.426, 0.416, 0.407, 0.4 , 0.388, 0.389, 0.387, 0.387, 0.373,
0.368, 0.363, 0.362, 0.36 , 0.359, 0.361, 0.373, 0.395, 0.425,
0.443, 0.446, 0.449, 0.449, 0.45 , 0.45 , 0.448, 0.455, 0.454,
0.464, 0.485, 0.63 , 0.753, 0.873, 0.84 , 0.793, 0.754, 0.764,
0.741, 0.544, 0.496, 0.413, 0.337, 0.341, 0.346, 0.378, 0.435,
0.468, 0.514, 0.554, 0.484, 0.42 , 0.447, 0.554, 0.639, 0.747,
0.991, 1.174, 1.382, 1.472, 1.527, 1.52 , 1.521, 1.523, 1.626,
1.694, 1.718, 1.778, 1.815, 1.937, 2.004, 2.018, 2.126, 2.203,
2.284, 2.384, 2.454, 2.49 , 2.514, 2.506, 2.464, 2.436, 2.413,
2.368, 2.369, 2.534, 2.579, 2.572, 2.583, 2.598, 2.618, 2.629,
2.648, 2.669, 2.676, 2.682, 2.657, 2.641, 2.576, 2.503, 2.447,
2.428, 2.447, 2.501, 2.546, 2.574, 2.743, 2.776, 2.754, 2.794,
2.799, 2.8 , 2.802, 2.806, 2.805, 2.805, 2.788, 2.754, 2.762,
2.779, 2.786, 2.792, 2.792, 2.793, 2.728, 2.486, 2.424, 2.386,
2.353, 2.304, 2.302, 2.268, 2.221, 2.206, 2.189], dtype=float32),
array([34.233, 34.24 , 34.243, 34.24 , 34.239, 34.241, 34.248, 34.246,
34.251, 34.254, 34.255, 34.262, 34.265, 34.271, 34.274, 34.276,
34.277, 34.279, 34.282, 34.284, 34.287, 34.289, 34.29 , 34.293,
34.297, 34.302, 34.305, 34.311, 34.315, 34.319, 34.321, 34.321,
34.321, 34.325, 34.329, 34.33 , 34.33 , 34.328, 34.334, 34.341,
34.343, 34.344, 34.346, 34.35 , 34.353, 34.358, 34.369, 34.378,
34.379, 34.38 , 34.398, 34.413, 34.415, 34.415, 34.411, 34.401,
34.385, 34.383, 34.386, 34.387, 34.389, 34.399, 34.403, 34.404,
34.419, 34.42 , 34.42 , 34.419, 34.422, 34.421, 34.422, 34.422,
34.423, 34.422, 34.424, 34.426, 34.429, 34.431, 34.43 , 34.427,
34.425, 34.425, 34.425, 34.425, 34.428, 34.433, 34.437, 34.441,
34.447, 34.453, 34.474, 34.475, 34.474, 34.488, 34.499, 34.507,
34.506, 34.504, 34.497, 34.486, 34.48 , 34.481, 34.488, 34.494,
34.495, 34.5 , 34.503, 34.501, 34.503, 34.512, 34.518, 34.516,
34.514, 34.511, 34.505, 34.502, 34.501, 34.502, 34.502, 34.502,
34.502, 34.501, 34.5 , 34.5 , 34.504, 34.502, 34.501, 34.501,
34.502, 34.502, 34.503, 34.505, 34.506, 34.508, 34.511, 34.512,
34.513, 34.514, 34.515, 34.518, 34.519, 34.521, 34.526, 34.532,
34.536, 34.537, 34.537, 34.537, 34.538, 34.538, 34.538, 34.538,
34.539, 34.54 , 34.543, 34.56 , 34.573, 34.587, 34.587, 34.584,
34.58 , 34.58 , 34.578, 34.563, 34.558, 34.553, 34.548, 34.548,
34.55 , 34.558, 34.569, 34.572, 34.578, 34.584, 34.58 , 34.574,
34.577, 34.588, 34.597, 34.608, 34.633, 34.653, 34.677, 34.688,
34.695, 34.694, 34.693, 34.692, 34.705, 34.714, 34.718, 34.725,
34.729, 34.743, 34.752, 34.754, 34.768, 34.778, 34.787, 34.8 ,
34.809, 34.814, 34.818, 34.818, 34.814, 34.81 , 34.808, 34.803,
34.804, 34.825, 34.832, 34.832, 34.833, 34.836, 34.838, 34.839,
34.842, 34.845, 34.846, 34.847, 34.846, 34.844, 34.838, 34.831,
34.825, 34.823, 34.824, 34.831, 34.838, 34.842, 34.864, 34.869,
34.867, 34.872, 34.873, 34.873, 34.873, 34.873, 34.873, 34.873,
34.871, 34.867, 34.868, 34.87 , 34.871, 34.871, 34.871, 34.872,
34.866, 34.84 , 34.833, 34.834, 34.827, 34.824, 34.821, 34.821,
34.816, 34.813, 34.812], dtype=float32))
Or into a pandas dataframe that we can export as a CSV or XLSX file for example
df = xrds[['TEMP','PSAL']].to_dataframe()
#df.to_csv('/path/to/file.csv')
df
| TEMP | PSAL | |
|---|---|---|
| PRES | ||
| 2.0 | 3.226 | 34.233002 |
| 3.0 | 3.211 | 34.240002 |
| 4.0 | 3.202 | 34.243000 |
| 5.0 | 3.205 | 34.240002 |
| 6.0 | 3.213 | 34.238998 |
| ... | ... | ... |
| 256.0 | 2.302 | 34.820999 |
| 257.0 | 2.268 | 34.820999 |
| 258.0 | 2.221 | 34.816002 |
| 259.0 | 2.206 | 34.813000 |
| 260.0 | 2.189 | 34.812000 |
259 rows × 2 columns
Looping through multiple files#
Now let’s look at how we can easily loop through all files (depth profiles) from one cruise
from siphon.catalog import TDSCatalog
base_url = 'https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708'
# Path to the catalog we can loop through
catalog_url = base_url + '/catalog.xml'
# Access the THREDDS catalog
catalog = TDSCatalog(catalog_url)
# Traverse through the catalog and print a list of the NetCDF files
catalog.datasets
['CTD_station_ISG_SVR1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_4_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG02_5_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG03_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG05_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG05_01_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG05_02_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG06_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG06_01_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG06_02_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG07_01_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG08_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG08_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG09_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG09_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG10_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG10_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG11_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG11_2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG12_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG14_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG15_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG16_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG17_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG18_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG19_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG20_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG22_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG23_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_NLEG24_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P1_NLEG01-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P1_NLEG01-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P1_NLEG01-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P2_NLEG04-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P2_NLEG04-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P2_NLEG04-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P2_NLEG04-4_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P3_NLEG07-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P3_NLEG07-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P3_NLEG07-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P3_NLEG07-4_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P3_NLEG07-5_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P4_NLEG11-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P4_NLEG11-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P4_NLEG11-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13-4_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13-5_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P5_NLEG13_1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P6_NLEG21_NPAL15-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P6_NLEG21_NPAL15-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P6_NLEG21_NPAL15-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P6_NLEG21_NPAL15-4_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P7_NLEG25_NPAL16-1_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P7_NLEG25_NPAL16-2_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc', 'CTD_station_P7_NLEG25_NPAL16-3_-_Nansen_Legacy_Cruise_-_2021_Joint_Cruise_2-1.nc']
Now let’s loop through this list and open each file in turn and print time_coverage_start attribute.
base_url = 'https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708'
catalog_url = base_url + '/catalog.xml'
catalog = TDSCatalog(catalog_url)
for dataset in catalog.datasets:
profile_url = base_url + '/' + dataset
xrds = xr.open_dataset(profile_url)
print(xrds.attrs['time_coverage_start'])
2021-07-12T19:05:04Z
2021-07-14T23:59:51Z
2021-07-15T02:01:30Z
2021-07-15T03:35:31Z
2021-07-15T04:45:26Z
2021-07-15T05:55:33Z
2021-07-15T07:25:12Z
2021-07-15T08:44:11Z
2021-07-16T17:22:02Z
2021-07-16T20:34:00Z
Combining data from all the files into a CSV or XLSX file#
import pandas as pd
base_url = 'https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708'
catalog_url = base_url + '/catalog.xml'
catalog = TDSCatalog(catalog_url)
# Initialize an empty list to store individual DataFrames
dataframes_list = []
for dataset in catalog.datasets:
profile_url = base_url + '/' + dataset
xrds = xr.open_dataset(profile_url)
profile_df = xrds[['TEMP','PSAL']].to_dataframe()
# Let's add some more columns from the global attributes that will help
profile_df['latitude'] = xrds.attrs['geospatial_lat_min']
profile_df['longitude'] = xrds.attrs['geospatial_lon_min']
profile_df['timestamp'] = xrds.attrs['time_coverage_start']
# Append the current DataFrame to the list
dataframes_list.append(profile_df)
# Concatenate all DataFrames in the list into a master DataFrame
master_df = pd.concat(dataframes_list)
# Reset index of the master DataFrame
master_df.reset_index(inplace=True)
master_df
#master_df.to_csv('/path/to/file.csv', index=False)
#master_df.to_excel('/path/to/file.csv', index=False) # Set index=False to exclude the index from being written to the Excel file
| PRES | TEMP | PSAL | latitude | longitude | timestamp | |
|---|---|---|---|---|---|---|
| 0 | 2.0 | 3.226 | 34.233002 | 78.128197 | 14.0032 | 2021-07-12T19:05:04Z |
| 1 | 3.0 | 3.211 | 34.240002 | 78.128197 | 14.0032 | 2021-07-12T19:05:04Z |
| 2 | 4.0 | 3.202 | 34.243000 | 78.128197 | 14.0032 | 2021-07-12T19:05:04Z |
| 3 | 5.0 | 3.205 | 34.240002 | 78.128197 | 14.0032 | 2021-07-12T19:05:04Z |
| 4 | 6.0 | 3.213 | 34.238998 | 78.128197 | 14.0032 | 2021-07-12T19:05:04Z |
| ... | ... | ... | ... | ... | ... | ... |
| 32028 | 3362.0 | -0.722 | 34.945000 | 82.003799 | 30.0427 | 2021-07-25T03:02:53Z |
| 32029 | 3363.0 | -0.722 | 34.945000 | 82.003799 | 30.0427 | 2021-07-25T03:02:53Z |
| 32030 | 3364.0 | -0.722 | 34.945000 | 82.003799 | 30.0427 | 2021-07-25T03:02:53Z |
| 32031 | 3365.0 | -0.722 | 34.945000 | 82.003799 | 30.0427 | 2021-07-25T03:02:53Z |
| 32032 | 3366.0 | -0.722 | 34.945000 | 82.003799 | 30.0427 | 2021-07-25T03:02:53Z |
32033 rows × 6 columns
Or maybe you prefer to have separate columns for each depth profile. Let’s create a dataframe with one pressure column and individual columns for the temperature data from each profile.
# Create an empty dictionary to store profile data
profile_data = {}
for dataset in catalog.datasets:
profile_url = base_url + '/' + dataset
xrds = xr.open_dataset(profile_url)
timestamp = xrds.attrs['time_coverage_start']
profile_data[timestamp] = xrds['TEMP'].to_dataframe()['TEMP']
# Creating a dataframe that includes all profiles
master_df = pd.DataFrame(profile_data.values()).transpose()
# Assigning the timestamps as column headers (currently TEMP for all)
master_df.columns = profile_data.keys()
# Display theb resulting DataFrame
master_df
| 2021-07-12T19:05:04Z | 2021-07-14T23:59:51Z | 2021-07-15T02:01:30Z | 2021-07-15T03:35:31Z | 2021-07-15T04:45:26Z | 2021-07-15T05:55:33Z | 2021-07-15T07:25:12Z | 2021-07-15T08:44:11Z | 2021-07-16T17:22:02Z | 2021-07-16T20:34:00Z | ... | 2021-07-19T14:29:47Z | 2021-07-20T13:58:18Z | 2021-07-20T19:58:26Z | 2021-07-21T17:35:04Z | 2021-07-22T13:57:51Z | 2021-07-23T06:12:20Z | 2021-07-23T09:25:55Z | 2021-07-24T01:04:37Z | 2021-07-24T14:31:40Z | 2021-07-25T03:02:53Z | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRES | |||||||||||||||||||||
| 2.0 | 3.226 | 3.782 | 3.035 | 2.504 | 1.960 | 1.735 | 1.694 | 1.966 | NaN | 1.382 | ... | NaN | 8.814 | NaN | -1.551 | -1.582 | -1.535 | -1.528 | NaN | -1.620 | -1.643 |
| 3.0 | 3.211 | 3.783 | 3.036 | 2.456 | 1.960 | 1.736 | 1.693 | 1.958 | 0.693 | 1.376 | ... | -1.499 | 5.513 | NaN | -1.560 | -1.585 | -1.541 | -1.524 | NaN | -1.630 | -1.643 |
| 4.0 | 3.202 | 3.783 | 3.036 | 2.456 | 1.950 | 1.735 | 1.691 | 1.954 | 0.662 | 1.362 | ... | -1.498 | 2.829 | NaN | -1.554 | -1.588 | -1.542 | -1.547 | NaN | -1.632 | -1.641 |
| 5.0 | 3.205 | 3.783 | 3.037 | 2.440 | 1.956 | 1.732 | 1.696 | 1.973 | 0.677 | 1.243 | ... | -1.497 | 0.875 | -0.452 | -1.552 | -1.588 | -1.549 | -1.550 | 1.913 | -1.632 | -1.639 |
| 6.0 | 3.213 | 3.792 | 3.035 | 2.443 | 1.961 | 1.733 | 1.695 | 1.976 | 0.708 | 1.359 | ... | -1.499 | 1.871 | -0.176 | -1.552 | -1.588 | -1.550 | -1.550 | 2.181 | -1.627 | -1.639 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3366.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | -0.722 |
| 3367.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3368.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3369.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3370.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
3370 rows × 62 columns
Maybe you want the coordinates as well as the timestamp in the column headers.
import pandas as pd
import xarray as xr
from siphon.catalog import TDSCatalog
base_url = 'https://opendap1.nodc.no/opendap/physics/point/cruise/nansen_legacy-single_profile/NMDC_Nansen-Legacy_PR_CT_58US_2021708'
catalog_url = base_url + '/catalog.xml'
catalog = TDSCatalog(catalog_url)
# Create an empty dictionary to store profile data
profile_data = {}
for dataset in catalog.datasets:
profile_url = base_url + '/' + dataset
xrds = xr.open_dataset(profile_url)
timestamp = xrds.attrs['time_coverage_start']
latitude = xrds.attrs['geospatial_lat_min']
longitude = xrds.attrs['geospatial_lon_min']
key = (timestamp, latitude, longitude)
profile_data[key] = xrds['TEMP'].to_dataframe()['TEMP']
# Create a MultiIndex DataFrame
index = pd.MultiIndex.from_tuples(profile_data.keys(), names=['timestamp', 'latitude', 'longitude'])
master_df = pd.DataFrame(profile_data.values(), index=index).transpose()
# Display the resulting DataFrame
master_df
| timestamp | 2021-07-12T19:05:04Z | 2021-07-14T23:59:51Z | 2021-07-15T02:01:30Z | 2021-07-15T03:35:31Z | 2021-07-15T04:45:26Z | 2021-07-15T05:55:33Z | 2021-07-15T07:25:12Z | 2021-07-15T08:44:11Z | 2021-07-16T17:22:02Z | 2021-07-16T20:34:00Z | ... | 2021-07-19T14:29:47Z | 2021-07-20T13:58:18Z | 2021-07-20T19:58:26Z | 2021-07-21T17:35:04Z | 2021-07-22T13:57:51Z | 2021-07-23T06:12:20Z | 2021-07-23T09:25:55Z | 2021-07-24T01:04:37Z | 2021-07-24T14:31:40Z | 2021-07-25T03:02:53Z |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| latitude | 78.128197 | 76.499802 | 76.595802 | 76.693001 | 76.764503 | 76.812202 | 76.914703 | 77.000198 | 78.000000 | 78.400002 | ... | 80.512199 | 80.480003 | 80.732498 | 81.547798 | 81.546700 | 81.542503 | 81.542702 | 82.000999 | 81.981697 | 82.003799 |
| longitude | 14.003200 | 31.219801 | 31.756800 | 32.303699 | 32.617298 | 32.897999 | 33.516300 | 34.002300 | 33.999699 | 33.999802 | ... | 33.814499 | 33.202499 | 33.121498 | 30.858700 | 30.795000 | 30.891500 | 30.846001 | 29.982201 | 29.979000 | 30.042700 |
| PRES | |||||||||||||||||||||
| 2.0 | 3.226 | 3.782 | 3.035 | 2.504 | 1.960 | 1.735 | 1.694 | 1.966 | NaN | 1.382 | ... | NaN | 8.814 | NaN | -1.551 | -1.582 | -1.535 | -1.528 | NaN | -1.620 | -1.643 |
| 3.0 | 3.211 | 3.783 | 3.036 | 2.456 | 1.960 | 1.736 | 1.693 | 1.958 | 0.693 | 1.376 | ... | -1.499 | 5.513 | NaN | -1.560 | -1.585 | -1.541 | -1.524 | NaN | -1.630 | -1.643 |
| 4.0 | 3.202 | 3.783 | 3.036 | 2.456 | 1.950 | 1.735 | 1.691 | 1.954 | 0.662 | 1.362 | ... | -1.498 | 2.829 | NaN | -1.554 | -1.588 | -1.542 | -1.547 | NaN | -1.632 | -1.641 |
| 5.0 | 3.205 | 3.783 | 3.037 | 2.440 | 1.956 | 1.732 | 1.696 | 1.973 | 0.677 | 1.243 | ... | -1.497 | 0.875 | -0.452 | -1.552 | -1.588 | -1.549 | -1.550 | 1.913 | -1.632 | -1.639 |
| 6.0 | 3.213 | 3.792 | 3.035 | 2.443 | 1.961 | 1.733 | 1.695 | 1.976 | 0.708 | 1.359 | ... | -1.499 | 1.871 | -0.176 | -1.552 | -1.588 | -1.550 | -1.550 | 2.181 | -1.627 | -1.639 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3366.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | -0.722 |
| 3367.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3368.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3369.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
| 3370.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | -0.719 | NaN | NaN |
3370 rows × 62 columns
Accessing a subset of the files#
We can use an if statement within our for loop to work with only certain files. Let’s say you are looking for data only within a certain longitude range. You can use select based on the relevant global attribute.
filtered_datasets = {}
for dataset in catalog.datasets:
profile_url = base_url + '/' + dataset
xrds = xr.open_dataset(profile_url)
longitude = xrds.attrs['geospatial_lon_min']
if 31 < longitude < 31.5:
filtered_datasets[round(float(xrds.attrs['geospatial_lat_min']),4)] = xrds
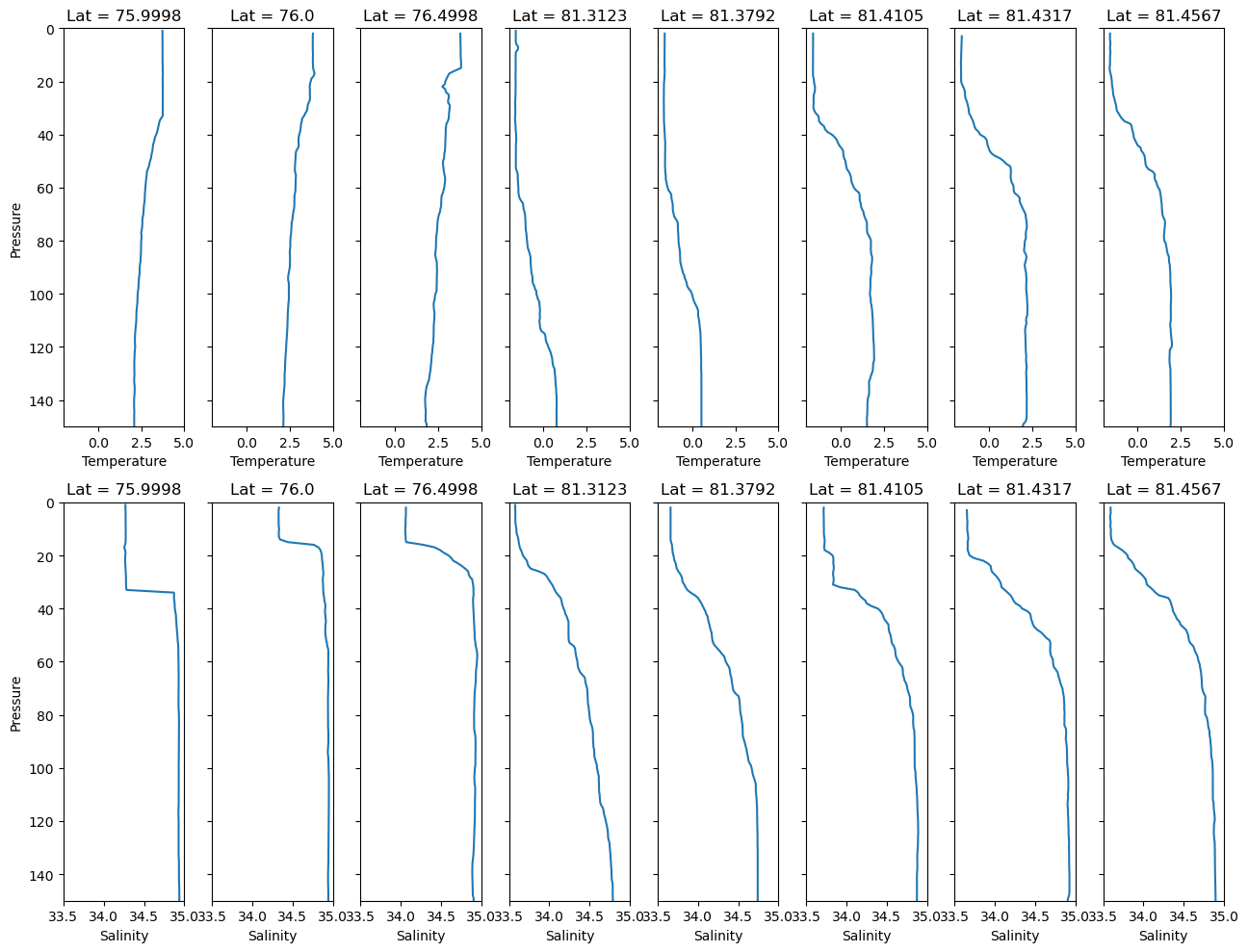
Plotting data from multiple files together#
How you do this is going to depend very much on the data and your requirements. However, this is a demonstration that it is possible to create a single plot of data from multiple NetCDF files.
import matplotlib.pyplot as plt
import xarray as xr
fig, axs = plt.subplots(2, len(filtered_datasets), figsize=(13, 10), sharey=True)
# Sorting the datasets in order of latitude
sorted_latitudes = sorted(filtered_datasets.keys())
for i, latitude in enumerate(sorted_latitudes):
xrds = filtered_datasets[latitude]
pressure = xrds['PRES']
temperature = xrds['TEMP']
salinity = xrds['PSAL']
axs[0,i].plot(temperature, pressure)
axs[0,i].set_title(f'Lat = {latitude}')
axs[0,i].set_ylim([150,0])
axs[0,i].set_xlim([-2,5])
axs[0,i].set_xlabel('Temperature')
if i == 0:
axs[0,i].set_ylabel('Pressure')
axs[1,i].plot(salinity, pressure)
axs[1,i].set_title(f'Lat = {latitude}')
axs[1,i].set_ylim([150,0])
axs[1,i].set_xlim([33.5,35])
axs[1,i].set_xlabel('Salinity')
if i == 0:
axs[1,i].set_ylabel('Pressure')
plt.tight_layout()
plt.show()